Description
Recombinant DNA techniques played a central role in the recent emergence of biology into the golden age. This program enables the advanced student to approach these frontiers in biotechnology. The program provides a challenging series of seven 3-hour laboratory sessions which are intended to give students hands-on-experience and a detailed understanding of these new investigative techniques and their potentials.
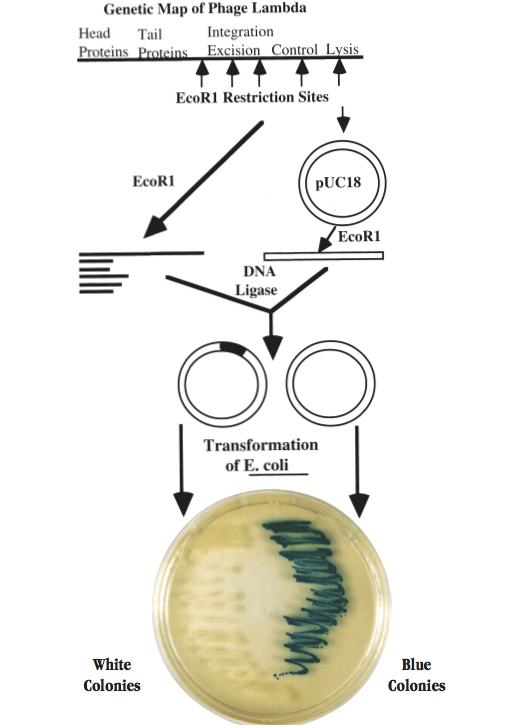
In the program, students prepare a library of recombinant plasmids which contain different regions of the lambda phage genome. They then amplify and characterize the lambda DNA in selected plasmids in order to identify the phage genes that they contain. The basic strategy followed in the program is shown on the facing page and is one that has been used by investigators around the world to clone DNA molecules from literally thousands of prokaryotic and eukaryotic organisms. However, many of the procedures have been modified to make them safe and suitable for a college laboratory that meets 3 hours once per week. The program provides essentially all of the instructions, chemicals, sterile media, and expendable accessories that are needed to carry out DNA electrophoresis, restriction nuclease digestions, DNA ligations, bacterial transformations, bacterial selection for ampicillin resistance and ß-galactosidase production, plasmid isolation, DNA hybridization, and Southern blot procedures. A centrifuge that can be operated at a force of at least 3,000 x g (such as a microcentrifuge or a larger floor model) and a water bath incubator that will maintain a temperature of 60-65°C are required for the program. A shaking bacterial incubator is strongly recommended.
Sample
A typical laboratory schedule for this program is given below where each lab session is approximately 3 hours.
Lab Session 1
Students digest phage lambda DNA with EcoR1 and ligate the resulting DNA fragments to EcoR1-digested pUC18 in order to create a series of recombinant plasmids. Some of the plasmids in the mixture will lack inserted lambda DNA while the others will have DNA inserts derived from various regions of the phage genome.
Lab Session 2
The recombinant molecules are characterized by electrophoresis and then introduced into competent E. coli cells by transformation. The cells are plated onto nutrient agar containing ampicillin and X gal. The ampicillin selects for all cells that have taken up plasmid while the X gal permits students to identify colonies which contain plasmids with lambda DNA inserts. The identification is based on a powerful color selection assay. Plasmid pUC18 contains a portion of the lac Z gene which codes for ß-galactosidase and X gal is a substrate of this enzyme. When pUC18 is carried by appropriate strains of E. coli, the lac Z gene is active and the ß-galactosidase converts X gal to a blue product which gives rise to blue colonies. If foreign DNA is inserted into the EcoR1 site of pUC18, the lac Z gene is inactivated and the colonies remain white.
Lab Session 3
Each student prepares overnight cultures from one blue colony which should lack lambda DNA, and three white colonies which should contain phage sequences. The plasmids are then isolated from the cultures by a unique mini-prep procedure that yields high quality plasmid DNA without the use of toxic organic solvents.
Lab Session 4
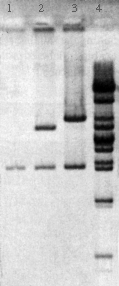
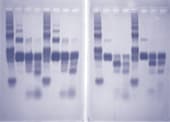
Students digest the plasmids with EcoR1, BamH1, and a combination of the two enzymes and determine the size of the DNA fragments by electrophoresis. The results of this analysis provide a first step in the identification of the lambda fragments that were cloned.
Lab Session 5 – 7
The DNA fragments in the gels are transferred to nylon membranes by the method of Southern using the transfer devices provided with the program. The fragments containing the lambda sequences are then detected by hybridization using a biotin-labeled probe made from phage lambda DNA. The results of this analysis and the restriction mapping studies performed during Lab Session 4 enable the student to identify the segment of the lambda genome in each plasmid.
In Early 2012 we updated the plasmid isolation procedure for this experiment. Revised Instructor manuals and the new Student pages are in a PDF format attached to the bottom of this page.
Revisions
In Early 2012 we updated the plasmid isolation procedure for this experiment. Revised Instructor manuals and the new Student pages are in a PDF format below:
Updated Instructor Guide
Updated Plasmid Isolation Procedure for S5 (4 pages)
 Due to Customs restrictions, we only accept orders from educational institutions within the Continental United States, Alaska or Hawaii.
Due to Customs restrictions, we only accept orders from educational institutions within the Continental United States, Alaska or Hawaii.